Statistical and computational methods for the microbiome
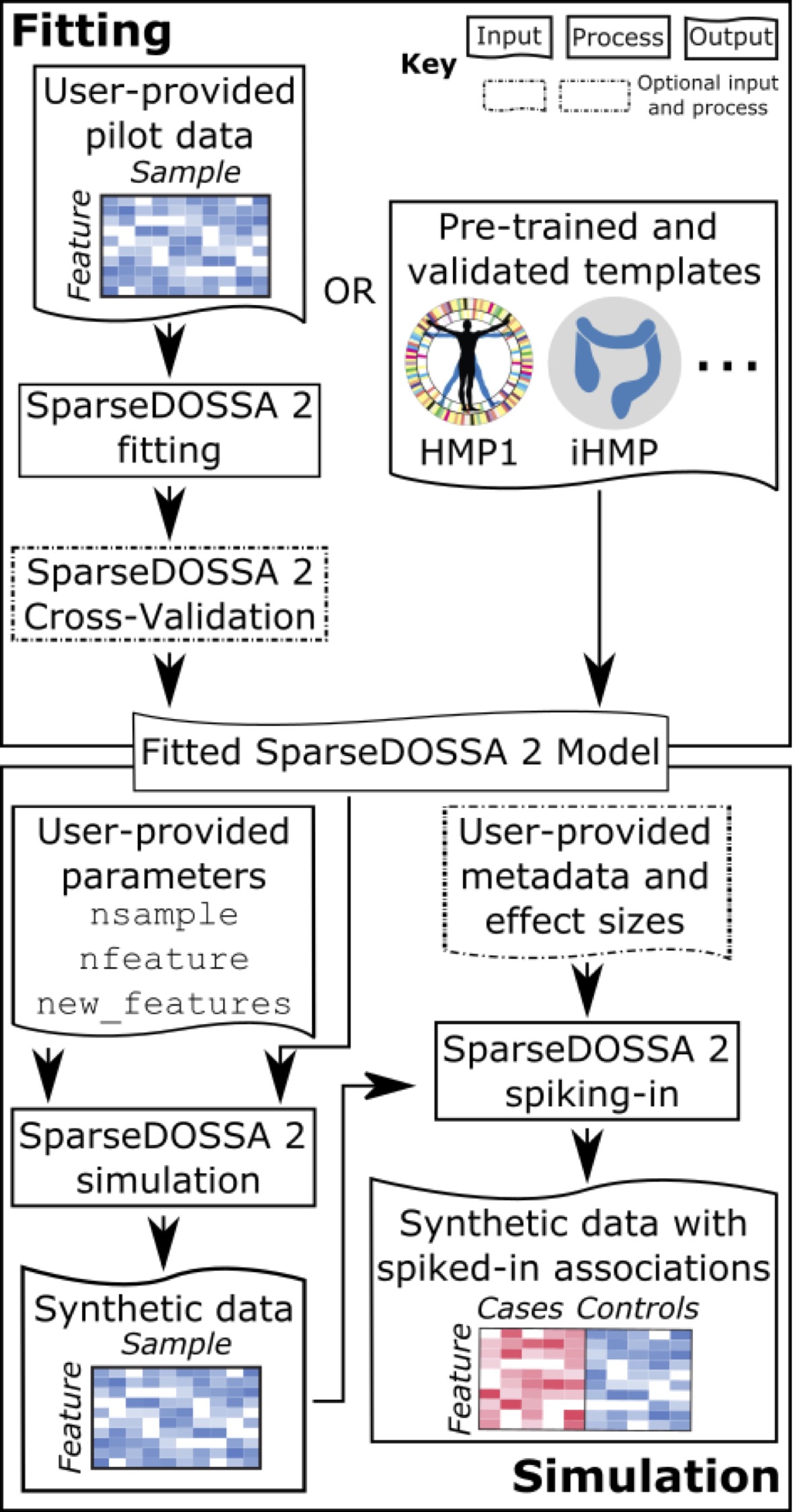
Common application problems in microbiome epidemiology include study design (power calculation), dimension reduction, biomarker discovery (association analysis), and data normalization and technical effect correction. Statistical methods, when targeting these applications, need to account for the unique characteristics of microbiome data, such as zero-inflation, compositionality, and high-dimensionality. These papers represent my efforts in this space (ordered by date of publication).
Ma, S. and Li, H., 2023. A tensor decomposition model for longitudinal microbiome studies. The Annals of Applied Statistics, 17(2), pp.1105-1126.
Ma, S. and Li, H., 2022. Statistical and Computational Methods for Microbial Strain Analysis. In Statistical Genomics (pp. 231-245). New York, NY: Springer US.
Ma, S., Shungin, D., Mallick, H., Schirmer, M., Nguyen, L.H., Kolde, R., Franzosa, E., Vlamakis, H., Xavier, R. and Huttenhower, C., 2022. Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using MMUPHin. Genome Biology, 23(1), pp.1-31.
Ma, S., Ren, B., Mallick, H., Moon, Y.S., Schwager, E., Maharjan, S., Tickle, T.L., Lu, Y., Carmody, R.N., Franzosa, E.A. and Janson, L., 2021. A statistical model for describing and simulating microbial community profiles. PLOS Computational Biology, 17(9), p.e1008913.
Mallick, H., Rahnavard, A., McIver, L.J., Ma, S., Zhang, Y., Nguyen, L.H., Tickle, T.L., Weingart, G., Ren, B., Schwager, E.H. and Chatterjee, S., 2021. Multivariable association discovery in population-scale meta-omics studies. PLOS Computational Biology, 17(11), p.e1009442.
Mallick, H., Ma, S., Franzosa, E.A., Vatanen, T., Morgan, X.C. and Huttenhower, C., 2017. Experimental design and quantitative analysis of microbial community multiomics. Genome Biology, 18(1), pp.1-16.
Collaborative microbiome research
Analyzing microbiome data in practice requires careful considerations for common issues and pitfalls, such as the approriate choice of quantitative method, interpretation of findings, and controlling false discovery findings. I am actively involved in collabroative, interdisciplinary research, to design the ideal analytical approach and ensure that scientific findings are robust and reproducible. These applications vary across different microbial ecology enivronments, disease phenotypes, and cohort designs.
Tan, Y., Shilts, M.H., Rosas-Salazar, C., Puri, V., Fedorova, N., Halpin, R.A., Ma, S., Anderson, L.J., Peebles Jr, R.S., Hartert, T.V. and Das, S.R., 2023. Influence of Sex on Respiratory Syncytial Virus Genotype Infection Frequency and Nasopharyngeal Microbiome. Journal of Virology, 97(3), pp.e01472-22.
Thompson, K.N., Oulhote, Y., Weihe, P., Wilkinson, J.E., Ma, S., Zhong, H., Li, J., Kristiansen, K., Huttenhower, C. and Grandjean, P., 2022. Effects of Lifetime Exposures to Environmental Contaminants on the Adult Gut Microbiome. Environmental Science & Technology, 56(23), pp.16985-16995.
Sinha, R., Abu-Ali, G., Vogtmann, E., Fodor, A.A., Ren, B., Amir, A., Schwager, E., Crabtree, J., Ma, S., Microbiome Quality Control Project Consortium and Abnet, C.C., 2017. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nature Biotechnology, 35(11), pp.1077-1086.
Spatial genomics
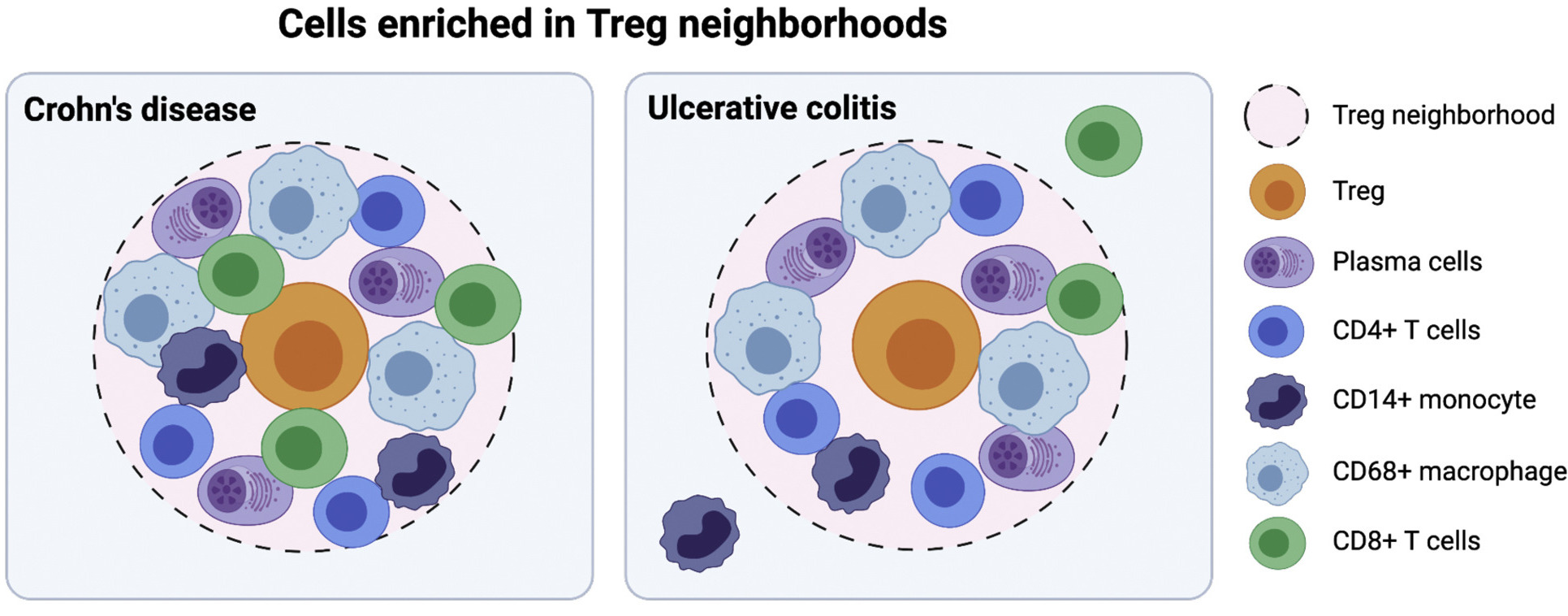
Spatial molecular assays (spatial transcriptome sequencing, multiplexed immunofluorescence imaging) provide crucial proximity information to study cellular and intercelluar activities in health and diseases. I am excited by this rapidly developing field and have made preliminary contributions, specifically for testing immune cell colocalization (paracrine cell-cell interactions).
Kondo, A., Ma, S., Lee, M.Y., Ortiz, V., Traum, D., Schug, J., Wilkins, B., Terry, N.A., Lee, H. and Kaestner, K.H., 2021. Highly multiplexed image analysis of intestinal tissue sections in patients with inflammatory bowel disease. Gastroenterology, 161(6), pp.1940-1952.
General computational genomics research
I have broad interests in applying quantitative methods to characterize the molecular profiles of various diseases. See examples below for studying the heterogenity of colorectal cancer and the dynamic cycle of malaria infection.
Ma, S., Ogino, S., Parsana, P., Nishihara, R., Qian, Z., Shen, J., Mima, K., Masugi, Y., Cao, Y., Nowak, J.A. and Shima, K., 2018. Continuity of transcriptomes among colorectal cancer subtypes based on meta-analysis. Genome Biology, 19(1), pp.1-14.
Dantzler, K.W., Ma, S., Ngotho, P., Stone, W.J., Tao, D., Rijpma, S., De Niz, M., Nilsson Bark, S.K., Jore, M.M., Raaijmakers, T.K. and Early, A.M., 2019. Naturally acquired immunity against immature Plasmodium falciparum gametocytes. Science Translational Medicine, 11(495), p.eaav3963.
Obaldia III, N., Meibalan, E., Sa, J.M., Ma, S., Clark, M.A., Mejia, P., Moraes Barros, R.R., Otero, W., Ferreira, M.U., Mitchell, J.R. and Milner, D.A., 2018. Bone marrow is a major parasite reservoir in Plasmodium vivax infection. MBio, 9(3), pp.e00625-18.
De Niz, M., Meibalan, E., Mejia, P., Ma, S., Brancucci, N.M., Agop-Nersesian, C., Mandt, R., Ngotho, P., Hughes, K.R., Waters, A.P. and Huttenhower, C., 2018. Plasmodium gametocytes display homing and vascular transmigration in the host bone marrow. Science Advances, 4(5), p.eaat3775.
Pelle, K.G., Oh, K., Buchholz, K., Narasimhan, V., Joice, R., Milner, D.A., Brancucci, N.M., Ma, S., Voss, T.S., Ketman, K. and Seydel, K.B., 2015. Transcriptional profiling defines dynamics of parasite tissue sequestration during malaria infection. Genome Medicine, 7, pp.1-20.